
Charted Waters
From the endangered Atlantic sturgeon to the elusive great white shark, New Jersey’s coastal waters are home to an array of species whose presence remains largely undetected unless spotted with the naked eye, or caught with fishing lines and trawl nets.
But now, thanks to advances in environmental DNA (eDNA) analysis, researchers like Assistant Professor of Biology Keith Dunton and his students can easily determine the presence, absence, and migration patterns of any native, invasive, endangered, or hard-to-find species in and around New Jersey’s coastal water bodies, just by dipping a cup into water.
“eDNA is a rapidly developing tool that I can foresee being utilized in the future for doing full species analysis,” says Dunton, who studies the sharks and sturgeon native to New Jersey.
Like humans, all animals shed DNA into their environment. Under water, this DNA might come from the slimy mucous layer that is continuously shed by an eel, or the excrement discharged by a shark. Undetectable to the eye, these small fragments of DNA float in the water and can be easily captured by dipping a regular-sized bottle into a body of water. Once filled, the bottles are brought back to campus, where researchers and their students analyze the eDNA captured (see sidebar, below).
The amount of DNA shed varies by species, but scientists recently determined that DNA remains in the water up to 48 hours, meaning even after the animal is long gone, researchers can tell they were there.
This ability to determine the presence of a particular species without having to physically spot or capture it is one of the myriad reasons why researchers believe eDNA could play a huge role in conservation biology.
For starters, eDNA collection is non-invasive, so it does not harm or traumatize the species being surveyed—which is particularly beneficial when studying critically endangered species. It’s also relatively low cost, and the water sampling process can be carried out by nearly anyone, which could potentially allow for a citizen science component in the future.
Another important aspect is that eDNA can be easily collected in various weather conditions and from places typically difficult to reach, allowing for easier and more continuous surveying of a species. That makes eDNA a perfect tool for researching a hard-to-catch fish such as the sea bass, which often lives near underwater structures and rocky environments.
While eDNA analysis has been around in some form since the 1980s, recent advancements in sequencing technology have made it cheaper, easier, and faster to use.
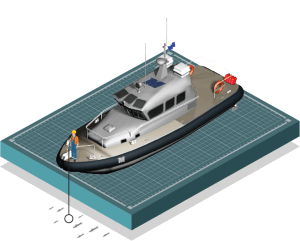
Professors Keith Dunton and Jason Adolf and their students use sterilized, clean water bottles to obtain samples from local waterways. One cup of water potentially contains the eDNA of dozens of species.

Back on campus, the students filter the seawater to collect eDNA contained in particles bigger than a half a micron, including microorganisms, detached cells of large organisms, and even free DNA.

Professor Megan Phifer-Rixey and her molecular biology students extract the eDNA on the filters and send them off to Rockefeller University, Monmouth’s partner university.

Researchers select primers, or short RNA strands, to target a specific type of organism group (e.g., vertebrate) or species (e.g., sturgeon). Then they “amplify” it using polymerase chain reaction (PCR)—think of it as making many photocopies of an area of DNA to make its signal more abundant. In the near future, amplification will be performed by Phifer-Rixey’s team on campus.
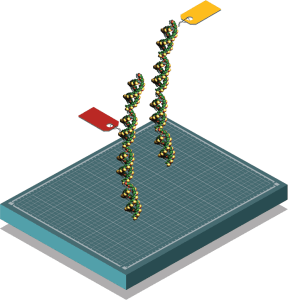
Unique “tags” are added to the amplified regions of DNA that identify where and when the sample was taken. In the near future, metabarcoding will be performed by Phifer-Rixey’s team on campus.

The tagged regions of DNA are sent to labs where they are sequenced in large batches on a Next Generation Sequencer, which spits out various series of unique, sequenced codes of DNA base pairs (ACTG) that help identify a specific species.

These sequenced codes can then run through a database that matches them to a species. This information is shared in databases that can be accessed by researchers nationwide.
Infographic by Don Foley
Dunton says the usefulness of eDNA testing became particularly popular in the early 2010s, when scientists used it to track the movements of an invasive fish species called silver carp. The carp, purposefully introduced to North America in the 1970s to control algae growth, wound up devastating native fish species reliant on that same algae growth. Scientists used eDNA to track the carp’s migration northward and develop a plan of where to focus resources in order to keep the fish from entering the Great Lakes.
“It’s a similar situation with sea lampreys,” Dunton says of the eel-like species of fish invading the Great Lakes and other water systems in North America. “For example, if you were investing money with lamprey control in the Great Lakes, you can sample a stream and say, ‘OK, lamprey aren’t here, so don’t put chemicals or resources into this stream.’”
In addition to determining species presence and migration patterns, eDNA sampling can also help scientists characterize an entire body of water to see how the species in it fluctuate and adapt to something as natural as the seasons changing, or as unnatural as an oil spill.
This summer, Jason Adolf, an endowed associate professor of marine science who studies harmful algae blooms, will be collecting eDNA samples from Sandy Hook Bay and the Shrewsbury/Navesink River systems in New Jersey as part of a long-term study of these systems.
While the benefits of eDNA analysis are expanding, the process isn’t without its limitations. For one, it cannot help researchers determine species abundance. It also cannot differentiate between the eDNA from a living animal and one that just died.
Because of this dilemma, the race is on amongst scientists to figure out how to expand the use of this new technology beyond detection of a species.
Since earlier this year, Dunton, Adolf, and researchers at the Rockefeller University, Monmouth’s partner university in eDNA analysis since 2016, have been trying to determine if there is a way to correlate eDNA results with those of the tried-and-true trawling methods traditionally used by biologists for conservation and management purposes.
For a full year, under the Urban Coast Institute–funded project, they are partnering with the New Jersey Department of Environmental Protection (NJDEP) to compare its trawl data—which includes information like the weight, length, and abundance of various species captured along the Jersey Shore—with eDNA results taken at the same time.
Already, the researchers have found that there are certain species detected only in the net, certain species primarily detected by eDNA, and species that show similar results via both methods.
“We’re seeing things with eDNA results that don’t normally get picked up in the trawl, like sea lamprey—which means they’re around, maybe attached to a fish, but we didn’t physically catch them,” Dunton says. “We also found all sorts of whale eDNA. You can see a whale surface but can’t trawl up a whale—luckily, their eDNA is abundant enough in the water to detect.”
With eDNA, scientists also have to determine how to rule out potential false positives. Mark Stoeckle, a senior research associate at Rockefeller University, got a positive hit for pig eDNA from a water sample taken in a Central Park lake—possibly the result of a pigeon or human dropping a small piece of ham or bacon into the water. Researchers also need to figure out a way to control for things like sewage contamination of water bodies, or cross-contamination in a lab.
This past fall, about 100 of the world’s leading eDNA researchers discussed these challenges and future possibilities for the science in New York City at the first National Conference on Marine Environmental DNA, co-hosted by Rockefeller and Monmouth’s Urban Coast Institute.
As Monmouth researchers continue to collect data in the field all across New Jersey and the Greater New York City area, that data, once analyzed, will be placed in a genetic reference library utilized by researchers throughout the country.
This supports fellow researchers in a speedy identification process as they analyze samples and try to make sense of large sets of data that support species-specific conservation efforts, invasive species control, and the greater understanding of whole ecosystems.
“In the future, eDNA analysis of marine waters will be an additional and indispensable tool in the marine ecologist’s toolbox that allows us to understand the fundamental question of ‘who’s out there’ in ocean ecosystems,” Adolf says.